| Biography | Research | Publications | Teaching | Group | Projects | Software | Sponsors | Collaborators |
|
|||||
|
Research Interests Computational Biology, Geometric Modeling, Image Processing, Computational Geometry, Computer Graphics, Compression, Mesh Generation, Scientific Computation, and Visualization My research areas of interest include Image Processing, Computational Geometry, Geometric Modeling, Computer Graphics, Visualization, and Computational Mathematics. Current research topics include the design and development of efficient and robust 2D/3D/4D image and geometry filtering, reconstruction, compression, matching and meshing algorithms. I am applying these algorithms to the structure elucidation and construction of multi-scale domain models of molecules, organelles, cells, tissues and organs from multi-modal, microscopy and bio-imaging. I am also involved in developing integrated approaches to computational modeling, mathematical analysis and interrogative visualization, especially for dynamic bio-medical phenomena. My research is currently funded by grants from the National Science Foundation (NSF) and the National Institutes of Health (NIH), and include
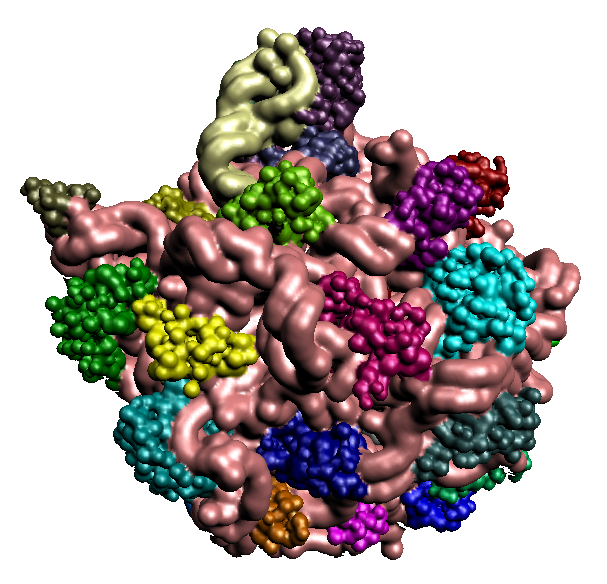
An interdisciplinary collaboration with Prof. Wah Chiu of Baylor College of Medicine, and Prof. Andrej Sali of University of California, San Francisco, to develop computational and visualization tools for feature extraction and structure modeling of large macromolecular complexes based on sequence data and in conjunction with subnanometer resolution cryo-Electron Microscopy (cryo-EM).
To develop and implement efficient algorithms for determining structural features of macromolecules from 3D-EM (Electron Microscopy) maps at multiple resolutions, and for generating hierarchical, volumetric spline approximations of the determined structural features to facilitate fast Fourier based matching of geometry and imaging.
The principal aims are to develop, implement and test novel mathematical algorithms that speed up computational protein-protein docking especially for larger problems, as well as to significantly improve the prediction of protein-protein binding. This collaborative project also has a subcontract to Dr. Art Olson and Dr. Michel Sanner at The Scripps Research Institute, San Diego, CA for testing and validation.
An interdisciplinary collaboration with Prof. J.T.Oden of The Institute for Computational Engineering and Sciences Prof. J.C.Browne of Computer Sciences , Prof. K.R.Diller of Biomedical Engineering , Dr. J.Hazle, MD of The University of Texas MD Anderson Cancer Center, to develop a dynamic data-driven planning, control, and visualization system for laser treatment of cancer. The proposed research is to develop a dynamic data-driven planning, control, and visualization system for the laser treatment of cancer. The proposed research includes development of a family of mathematical and computational models of hio-heat transfer, tissue damage, and tumor viability, dynamic calibration, verification and validation processes based protocols using model predictions.
|
|||||